Welcome to spann's documentation!
SPANN
paper “SPANN: Annotating
single-cell resolution spatial transcriptome data with scRNA-seq data”
(under peer-review at bib) 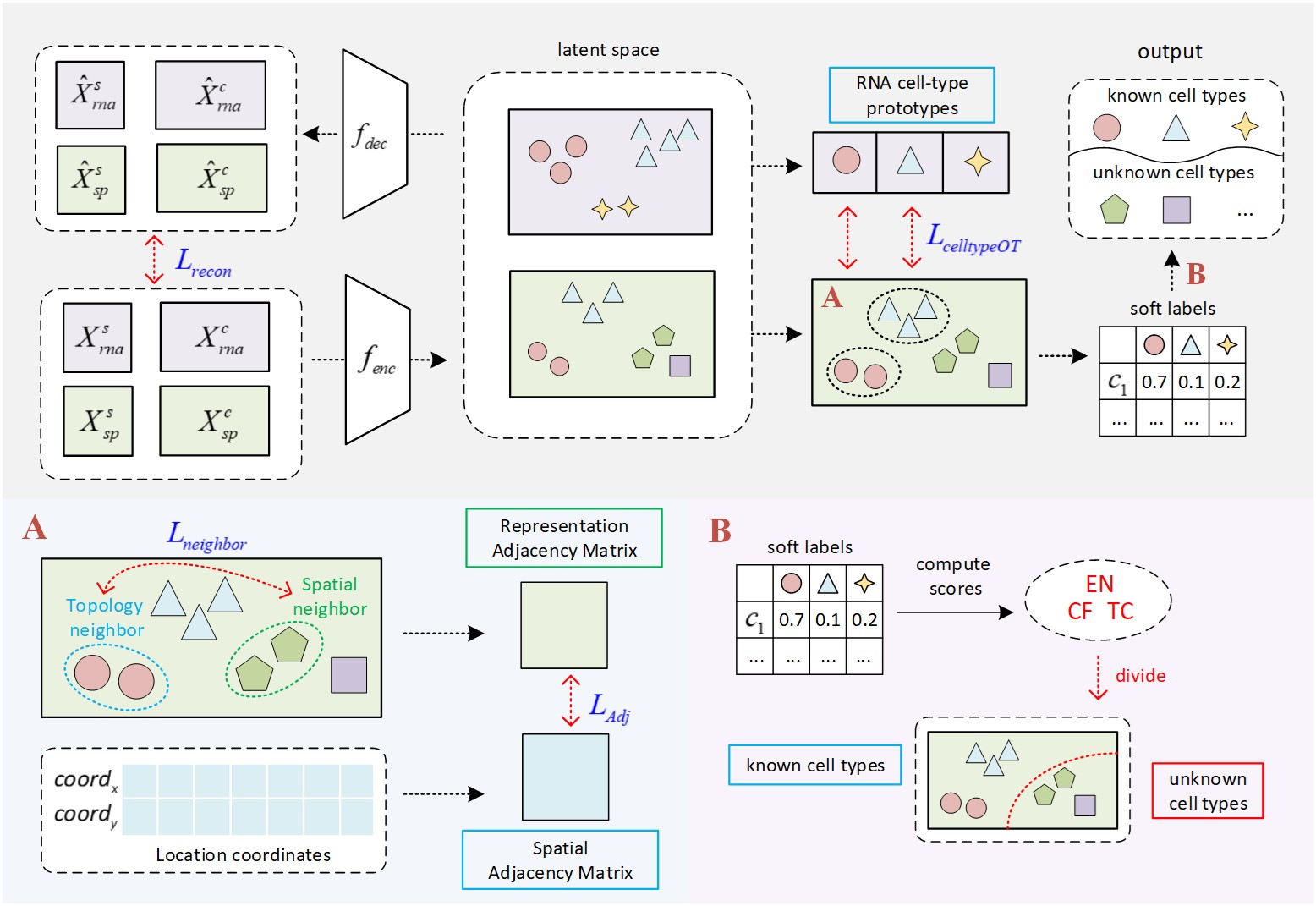
File Descriptions and data requirement
SPANN is a single-cell resolution spatial transcriptome data annotator. With a well-annotated reference scRNA-seq data, one can accurately identify cell identifications as well as discover novel cells.
There are 2 tutorials in the repository. Tutorial 1 annotates spatial transcriptome data without validation. If you do not have cell type labels for spatial data, you can follow tutorial 1 to annotate the spatial data with scRNA-seq reference. Tutorial 2 annotates spatial transcriptome data and evaluate the annotation performance with ground truth cell type labels. If you want to validate the annotation performance of SPANN or compare SPANN to other methods on benchmark datasets, you can follow tutorial 2.
The Mouse Embryo seqFISH-scRNA datasets used in the tutorials can be downloaded from https://drive.google.com/drive/folders/1kEHvid7F43sZAWh4xB-P0gLgYEnXVnNS?usp=sharing
We also uploaded the notebooks applying SPANN on all datasets mentioned in our paper.
Hyper-parameters and recommended settings
lambda_recon: default 2000, the strength of reconstruction loss
lambda_kl: default 0.5, the strength of KL-divergence loss
lambda_cd: default 0.001, the strength of cell-type-level domain alignment loss, we recommend to set it higher(=0.1) when the gap between scRNA-seq data and spatial data is big, and to set it lower(=0.0001) when the gap between datasets is small.
lambda_spa: default 0.1, the strength of spatial-representation adjacency loss, we recommend to set it lower(=0.01) when the spatial pattern is not clear(when the spatial distribution is chaotic), and recommend to set it higher(=0.5) when the spatial pattern is clear.
lambda_nb: default 0.1, the strength of neighbor loss, we recommend to set it lower(=0.01) when the spatial pattern is not clear(when the spatial distribution is chaotic), and recommend to set it higher(=10) when the spatial pattern is clear.
mu: default 0.6, the update speeed of beta (from 0.0-1.0). beta is the estimate of target cell type distribution.
temp: default 0.1, the temperature paremeter for spatial-representation adjacency loss